A02-1 Modality of nucleosome dynamics
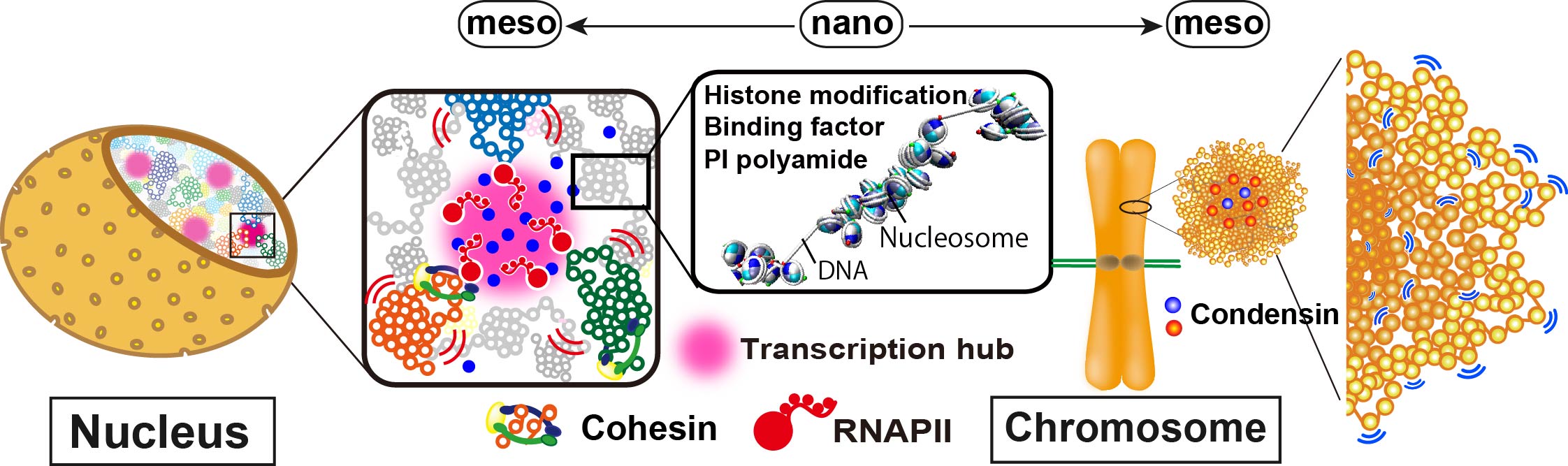
The purpose of this study is to understand “genome modality” from the viewpoint of the packing structure and dynamics of the nucleosome, which is the basic structural unit of genome chromatin. We have so far revealed that the local movement of nucleosomes, presumably caused by thermal fluctuations, is constrained by various protein complexes such as transcription machinery (left), cohesin (left), and condensin (right). The local movement of nucleosomes thus reflects the organization of chromatin in living cells and is directly linked to the understanding of “genomic modality.” In this study, to examine the movement of nucleosomes and their packing structure, we will use single-molecule imaging (Maeshima/Hibino), specific DNA sequence-binding polyamide/high-speed AFM (Sugiyama), and genome sequencing (Taniguchi). Besides, through collaborative research, we will investigate cells derived from patients with cohesin-related diseases. We contribute to understanding the abnormality of the chromatin organization, which induces various genetic diseases, as well as the organization of interphase chromatin (left) and mitotic chromosomes (right).

Kazuhiro Maeshima Project Leader
- Department of Chromosome Science, National Institute of Genetics
- http://maeshima-lab.sakuraweb.com

Kayo Hibino Co-Project Leader
- Department of Chromosome Science, National Institute of Genetics
- http://maeshima-lab.sakuraweb.com

Hiroshi Sugiyama Co-Project Leader
- Graduate School of Science, Kyoto University
- http://kuchem.kyoto-u.ac.jp/chembio/

Yuichi Taniguchi Co-Project Leader
- Institute for Advanced Study, Kyoto University
- https://www.icems.kyoto-u.ac.jp/ja/wwa/taniguchi/

